“Thanks to the power of open source, the compute capability provided by the HPE Spaceborne Computer-2, and the scalability of Azure, we are empowering developers to build for space at a speed that’s out of this world.”—Kevin Mack, Senior Software Engineer, Microsoft
This morning Microsoft News published a story about the use of Azure, enabled by HPE’s Spaceborne Computer-2 on the International Space Station (ISS). The project was designed to overcome the limited bandwidth between ISS and Earth by validating the benefits of a computational workflow that spans edge and cloud. Under this workflow, examination of high-volume raw data is processed and performed on the ISS using the HPE Spaceborne Computer-2’s edge computing platform and a much smaller data set containing only “interesting bits” is sent to Earth, where cloud resources are used to perform compute-intensive analysis to determine what those interesting bits really mean.
The Azure Space team performed the software development needed for the entire experiment in just three days.
A brief background
The International Space Station (ISS), a microgravity and space environment research laboratory, has just observed 20 years of continuous human presence. New technology is delivered to it regularly, as needed to keep up with the research being performed. Computers used on the ISS have typically been custom-built with specialized hardware and programming models, needed to deliver the reliability needed in space. Unfortunately, the developer experience for targeting these custom spaceborne systems is complex, making programming slow and challenging compared to the commercial-off-the-shelf systems used by most developers today.
Installed in 2017, Spaceborne Computer-1, designed by HPE, validated that a modern, commercial-off-the-shelf computer could survive a launch into space, be installed by astronauts, and operate correctly on the ISS—without “flipping bits” due to increased radiation in space. Basically, it was a year-long test to see if the computer hardware used on Earth would function normally in space. Building on this success, HPE’s Spaceborne Computer-2, an edge computing platform with purposely designed features for harsh environments, was installed in April 2021 to deliver twice as much compute performance, and for the first time, artificial intelligence (AI) capabilities to advance space exploration and research by enabling the same programming models and developer experiences used on Earth.
In many ways, Spaceborne Computer-2, which is comprised of the HPE Edgeline EL4000 Converged Edge system and HPE ProLiant DL360 Gen10 server, is the ultimate edge computing device platform, putting a game-changing amount of compute at the edge of space. However, the real limiting factor is the bandwidth between the ISS and Earth. Although Spaceborne Computer-2 supports the maximum available network speeds, it only receives from NASA an allocation of two hours of communication bandwidth a week to transmit data to earth, with a maximum download speed of 250 kilobytes per second.
In some cases, working around limited bandwidth can be accomplished by HPE helping researchers to compress data on Spaceborne Computer-2 before sending it down to Earth. In other cases, the data can be fully analyzed in space without needing to use the slow downlink at all. But what about research that requires more compute or bandwidth than what Spaceborne Computer-2 can provide, or that can be allotted to a single experiment among many? To address such scenarios, HPE applied its vision for an “edge to cloud” experience, in which Spaceborne Computer-2 is used to perform preliminary analysis or filtering on large data sets, extract what’s interesting or unexpected, and then burst those results down to Earth and into the public cloud for full analysis.
The Azure Space experiment
The Azure Space team at Microsoft proposed an experiment that simulates how NASA might monitor astronaut health in the presence of increased radiation exposure, as exists outside of our protective atmosphere. Such exposure will only increase as astronauts venture beyond the ISS’s low-earth orbit into and beyond the Van Allen Belts.
The experiment assumes access to a gene sequencer onboard the ISS, which is used to regularly monitor blood samples from astronauts. However, gene sequencing can generate an incredible amount of data—far too much for a 2Mbps/sec downlink—and the output needs to be compared against a large clinical database that’s constantly being updated.
To overcome those limitations, the experiment uses HPE Spaceborne Computer-2 to perform the initial process of comparing extracted gene sequences against reference DNA segments and capture only the differences, or mutations, which are then downloaded to the HPE ground station.
On earth, the data is uploaded to Azure, where the Microsoft Genomics service does the computational “alignment” work—the process of matching the short base-pair gene sequence reads in the downloaded data (which are about 70 base pairs in length) against the full 3 giga-base-pair human genome, as required to determine where in the human genome each mutation is located and the type of change (deletion, addition, replication, or swap). Aligned reads are then checked against the National Institute for Health’s dbSNP database to determine what the health impacts of a given mutation might mean. Watch the video below to see Azure in action.
Development process and computational workflow
The entire experiment was coded by 10 volunteers from the Azure Space team and its parent organization, the Azure Special Capabilities, Infrastructure, and Innovation Team. All major software components (both ISS-based and Azure-based) were written in Python and bash using Visual Studio Code, GitHub, and the Python libraries for Azure Functions and Azure Blob Storage. David Weinstein, Principal Software Engineering Manager at Azure Space, led the three-day development effort—consisting of a one-day hackathon and two days of cleanup.
The following graphic shows the computational workflow. It starts on the ISS, on Spaceborne Computer-2, which runs Red Hat Linux 7.4.
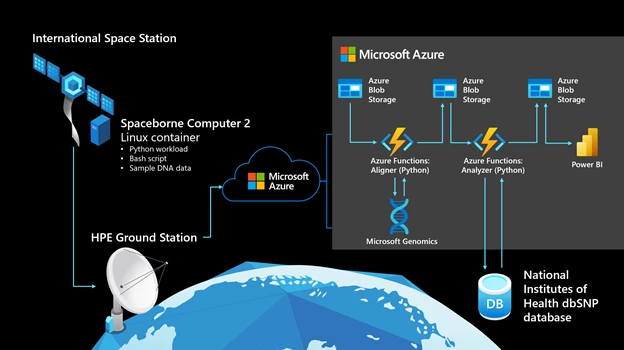
In space
- A Linux container hosts a Python workload, which is packaged with data representing mutated DNA fragments and wild-type (meaning normal or non-mutated) human DNA segments. There are 80 lines of Python code, with a 30-line bash script to execute the experiment.
- The Python workload generates a configurable amount of DNA sequences (mimicking gene sequencer reads, about 70 nucleotides long) from the mutated DNA fragment.
- The Python workload uses awk and grep to compare generated reads against the wild-type human genome segments.
- If a perfect match cannot be found for a read, it’s assumed to be a potential mutation and is compressed into an output folder on the Spaceborne Computer-2 network-attached storage device.
- After the Python workload completes, the compressed output folder is sent to the HPE ground station on Earth via rsync.
On Earth
- The HPE ground station uploads the data it receives to Azure, writing it to Azure Blob Storage through azcopy.
- An event-driven, serverless function written in Python and hosted in Azure Functions monitors Blob Storage, retrieving newly received data and sending it to the Microsoft Genomics service via its REST API.
- The Microsoft Genomics service, hosted on Azure, invokes a gene sequencing pipeline to “align” each read and determine where, how well, and how unambiguously it matches the full reference human genome. (The Microsoft Genomics service is a cloud implementation of the open-source Burroughs-Wheeler Aligner and Genome Analysis Toolkit, which Microsoft tuned for the cloud.)
- Aligned reads are written back to Blob Storage in Variant Call Format (VCF), a standard for describing variations from a reference genome.
- A second serverless function hosted in Azure Functions retrieves the VCF records, using the determined location of each mutation to query the dbSNP database hosted by the National Institute of Health—as needed to determine the clinical significance of the mutation—and writes that information to a JSON file in Blob Storage.
- Power BI retrieves the data containing clinical significance of the mutated genes from Blob Storage and displays it in an easily explorable format.
The Aligner and Analyzer functions total about 220 lines of code, with the Azure services and SDKs handling all of the low-level “plumbing” for the experiment. The functions are automatically triggered by blob storage uploads and are configured to point to the right storage accounts—requiring just a small amount of code to parse the raw data and query Microsoft Genomics and the dbSNP database at runtime.
Develop and test
During development and test, developers didn’t have access to HPE Spaceborne Computer-2 or the HPE ground station, so they recreated those environments on Azure, relying on GitHub Codespaces to further increase their velocity. They packaged both the ISS and ground station environments into an Azure Resource Manager (ARM) template, which simulates the latency between the ISS and the ground station by deploying the Spaceborne Computer-2 environment to an Azure data center in Australia and the ground station environment to one in Virginia.
The results
On August 12, 2021, the 120MB payload containing the experiment developed by Azure Space was uploaded to the ISS and run on Spaceborne Computer-2. The experiment is configurable, so Azure Space was able to execute “test”, “small”, and “medium” scenarios, executed in that order. One week later, after learning from the first run and optimizing processing on SBC-2, the team was able to run a full human genome size file on the ISS.
Table 1 shows the results of both experiments in terms of processing times and data volumes:
|
Test |
Small |
Medium |
Full human genome |
|
|
Raw data examined |
500KB |
6MB |
150MB |
182GB |
|
Downloaded to Earth |
4KB |
40KB |
900KB |
13MB |
|
Run time on ISS |
20 seconds |
2 minutes |
1 hour |
78 minutes |
|
Download time from ISS |
<1 second |
2 seconds |
17 seconds |
1:56 minutes |
The experiment’s successful completion—and the data collected through it—is proof of how an edge-to-cloud computing workflow can be used to support high-value use cases aboard the ISS that might otherwise be impossible due to compute and bandwidth constraints. Without preprocessing the simulated output of the gene sequencer on the ISS to filter out only the gene mutations, 150 times as much data would need to be downloaded to Earth. Thus, a 200GB raw full human genome read which would require over two years to download given bandwidth and downlink window constraints, was successfully filtered to 13MB—which was transmitted in just under two minutes.
Similarly, attempting to perform all of the processing that’s being done on Azure would require uploading a copy of the full reference human genome and a copy of the full dbSNP database. To complicate matters, the dbSNP database is constantly being updated and peer-reviewed by scientists across the globe, meaning that regular synchronization would be required to maintain a useful copy in space.
Build cloud applications productively, anywhere
From a software development perspective, the developer velocity with which Azure Space delivered the experiment is as impressive as its results—with all components delivered over a three-day period using serverless Azure Functions written in Python, and best-in-class developer tools such as Visual Studio Code and GitHub. To support the development of additional experiments by others, Weinstein’s team at Azure Space published the Resource Manager templates containing the simulated ISS and ground station environments they used for development and test.
Making such capabilities available to others is just one early step for Azure Space, a new vertical within Microsoft that was publicly announced about a year ago. Its twofold mission: to enable organizations who build, launch, and operate spacecraft and satellites and to “democratize the benefits of space” by enabling more opportunities for all actors, large and small, in much the same way that support for open source on Azure has helped democratize cloud computing. One such example is Azure Orbital, a ground station as-a-service that provides communication and control for satellite operators—including the ability to easily process satellite data at a cloud-scale.
Conclusion
Today, HPE’s Spaceborne Computer-2 is performing its mission of enabling “edge-to-cloud” experiments and proofs-of-concepts (POCs). Initially, these are fully isolated from the operational systems that astronauts and NASA depend on to operate the ISS itself.
As humankind continues to push farther into space, having real compute power at the edge will become more and more important. Use cases will increase, and developers who employ compute-intensive data science, AI, and machine learning tools will always be pushing the limits of the compute resources they have available to build the next generation of cloud-native applications. The edge-to-cloud compute model is one solution to such challenges, and the genomics experiment by Azure Space validates how that model can help “democratize space” for the benefit of all. As one of the developers on the Azure Space team put it, “Anyone could adapt what we built to their own needs because it’s all open source—based on tools and technologies that any high school programming class could download and use.”
Learn more about open-source development on Azure.

